Académie d'Excellence "Territoires, Environnements, Risques et Résilience"
Metabolomics for Exposure to Chemical or Radiological Risks
Academy 3 highlight
Metarisk brings together chemists, biologists, physicians, mathematicians, and risk management professionals to develop a new method to measure toxicological risks in a population and to help manage crisis situations.
The project
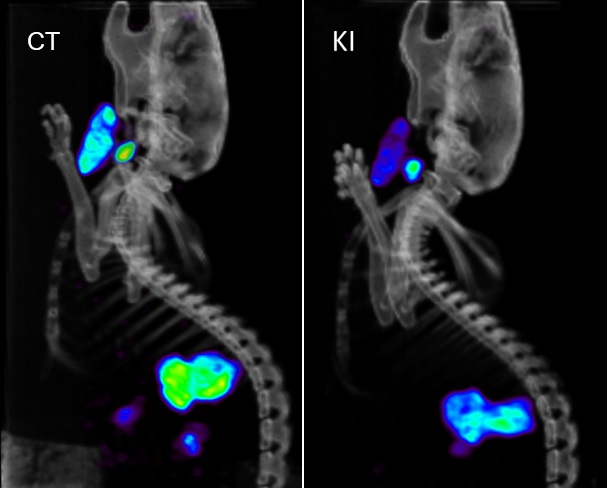
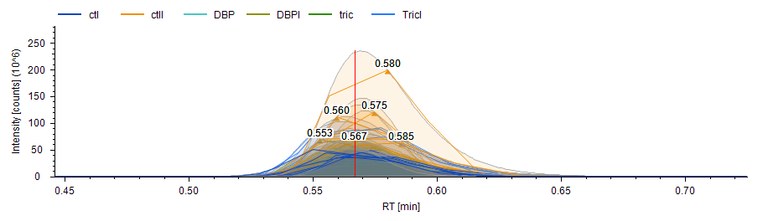
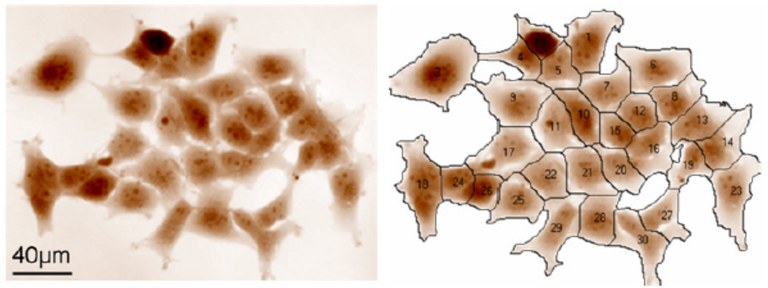
Metarisk aims to develop new metabolomics methods to assess a population’s exposure to chemical or radiotoxicological contaminations. Our strategy does not aim to identify exposure sources (such as toxic chemical agents) but, rather, to identify metabolomics biomarkers of exposure. Metabolomics yields qualitative and quantitative information on the nature and abundance of specific small organic molecules in biological samples that can attest to their contamination. The identification of these specific molecules should then enable discrimination between exposed and non-exposed individuals in a given population. The Metarisk project was launched recently: Our objective is to identify and characterize urinary metabolomic signatures of specific biological effects following exposure to different chemical sources. To best benefit from the combined expertise of the Metarisk team, we are initially focusing on chemical compounds that induce thyroid disruptor effects, radioactive iodine from nuclear industry accidents, and low-dose natural uranium. Meanwhile, we are developing methods for metabolomic analyses (LC-MS/MS) and for post-processing of metabolomics data (machine and deep learning). The work is in progress. We expect that the new metabolomics methods will become a useful tool for contamination tracking in a human population. As such, they would be helpful for population treatment, crisis management, and legal determination of individual exposure. The new metabolomics methods should also be useful for fundamental research in biology, toxicology and clinical research.
The +
If successful, this project should allow for the rapid identification of individuals who have been exposed to chemical or radiological contamination. These individuals could therefore be treated by health care professionals using appropriate crisis management procedures.
What’s next?
The capacity to detect and monitor biological effects of contamination using metabolomics should prove useful to optimize the management of related risks by crisis managers. To generalize and expand the scope of our results, we will build on the data obtained within the frame if this project to apply for additional funding from the European Union.
Project information
|
Scientific domain
Toxicology Exposure metrology
|
Key words
Metabolomics
Spectrometry Big data Machine learning |
||
|
Total budget
177.1 k€ including :
172.1 k€ from Académie 3 ; 5 k€ from Académie 4 |
Students inolved
Benjamin Reeves, PhD ICN |
||
| Partner laboratories
ICN
IPMC ESPACE INRIA CAL Géoazur GREDEG |
Project members
|
Project valorization
Publication :
- Primal-Dual for Classification with Rejection (PD-CR): A novel method for classification and feature selection. An application in Omics studies. Submitted to Computational and Structural Biotechnology Journal




















